Metadata
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is FAIR? What is the origin of the FAIR movement?
Why is FAIR important?
What is the difference between FAIRness and FAIRification of data?
Objectives
Define metadata and its various types
Recall the community standards and how to apply them to data and metadata
What is metadata?
Metadata refers to the information that describes your data.
In other words, imagine you have an Excel spreadsheet containing data values for an assay. You would use column headings to assign meaning and context. These column headings are your metadata, explaining the data values in each cell. In addition, any documentation or explanation of the accompanying excel file is also considered metadata.
Let’s look at Figure 1, showcasing a spreadsheet containing data for a clinical assay. In this example, the data are the patient ID, disease type, and heart rate values. The metadata, the column headings, describe that those values correspond to the patient ID, disease type, and heart rate, as well as the name of the cohort and the contact e-mail.

What information could we add to better understand the data contained in the dataset?
We could add additional metadata to indicate data provenance, i.e. data origin, what happens to it or where it moves over time. In this case, we should add more information about the cohort name. “Human Welsh Cohort” does not tell us much about the data if compared to other Welsh cohorts. Instead, we could include the following:
- A unique ID or detailed title for the cohort
- A project URL describing its origin and composition At first glance, the column headings seem descriptive. However, taking a closer look, we can see that the “disease type” column captures both the disease type and stage, which can cause ambiguity:
- In row 3, it is unclear whether the disease is diabetes mellitus or insipidus
- In row 4, it is unclear whether the type of diabetes mellitus is 1 or 2
- There is an empty space in the final row. It is unclear whether this is due to a lack of information or that the patient does not have diabetes
Instead, we can create two separate columns: one for disease type and one for the stage (see Figure 2).

The importance of metadata
Metadata is small and can be easily maintained not only in the database but in personal computers. The maintenance of datasets in a public database comes at a cost. It can be minimised when maintaining the metadata instead. In addition, metadata is also highly efficient for sharing sensitive data. The details available are those provided in the metadata, such contact details of the researchers, how to get the data and how it was generated.
Types of metadata
We’ve seen that metadata can describe various aspects of your dataset. Based on how to FAIR, there are three types of metadata:
- Descriptive Metadata: this metadata type help you identify the dataset e.g identifiers, publication name .. etc
- Structural Metadata describes how the dataset is generated and structured internally e.g. analysis, units of measurements, collection methods … etc
- Administrative metadata describes who was in charge of the data, who worked on the project, and how much money was spent.
Let’s look at an example using microarray data from the ArrayExpress database (Figure 3) to locate the different types of metadata that we have defined.
 We can observe:
We can observe:
- Administrative metadata: authors and organisations underneath the dataset title, and the information in “Publication”
- Descriptive metadata: “Description” section that sumarises the information contained in the dataset
- Structural Metadata: “Protocols”, “Samples” and “Assay and Data” sections describing the structure of the dataset and how it was generated
Metadata is also a documentation of data lifecycle. Through metadata, you describe what happened to metadata at each step of data life cycle. We use the term Data provenance to describe these steps. Provenance is the detailed description of the history of the data and how it is generated. Here is an example from arrayexpress database where there is accurate description of the microarray life cycle. As you can see in this example from E-MTAB-6980 dataset, there is rich description of the study design, organism, platform and timing of data collection.
How to use metadata to describe your dataset?
Metadata is data about data! It is important to know how to document it and use the right vocabulaeries to make your metadata FAIR. Using the right voacabularies will help standardize the way we describe our data.
Vocabularies and ontologies Controlled vocabularies: are list of terms that describes certain domain of knowledge. Vocabularies usually include definition of the term and any synonyms. For instance, medical subject headings (MeSH) terms is a common resource for controlled vocabularies. For instance, you can describe carotid artery as “common carotid artery” or “carotid sinus”
When you describe your data, you also need to describe the relationship between different vocabularies, which we call it ontologies
Ontologies: describe the relationship between different terms. There are many resources that you can use to get ontologies for your metadata. BRENDA, an ELIXIR resource that helps you get the right ontologies for your metadata.
Exercise
You are researcher working in the field of food safety and you are doing clinical trial, do you know how to > choose the right vocabularies and ontologies for it?
solution
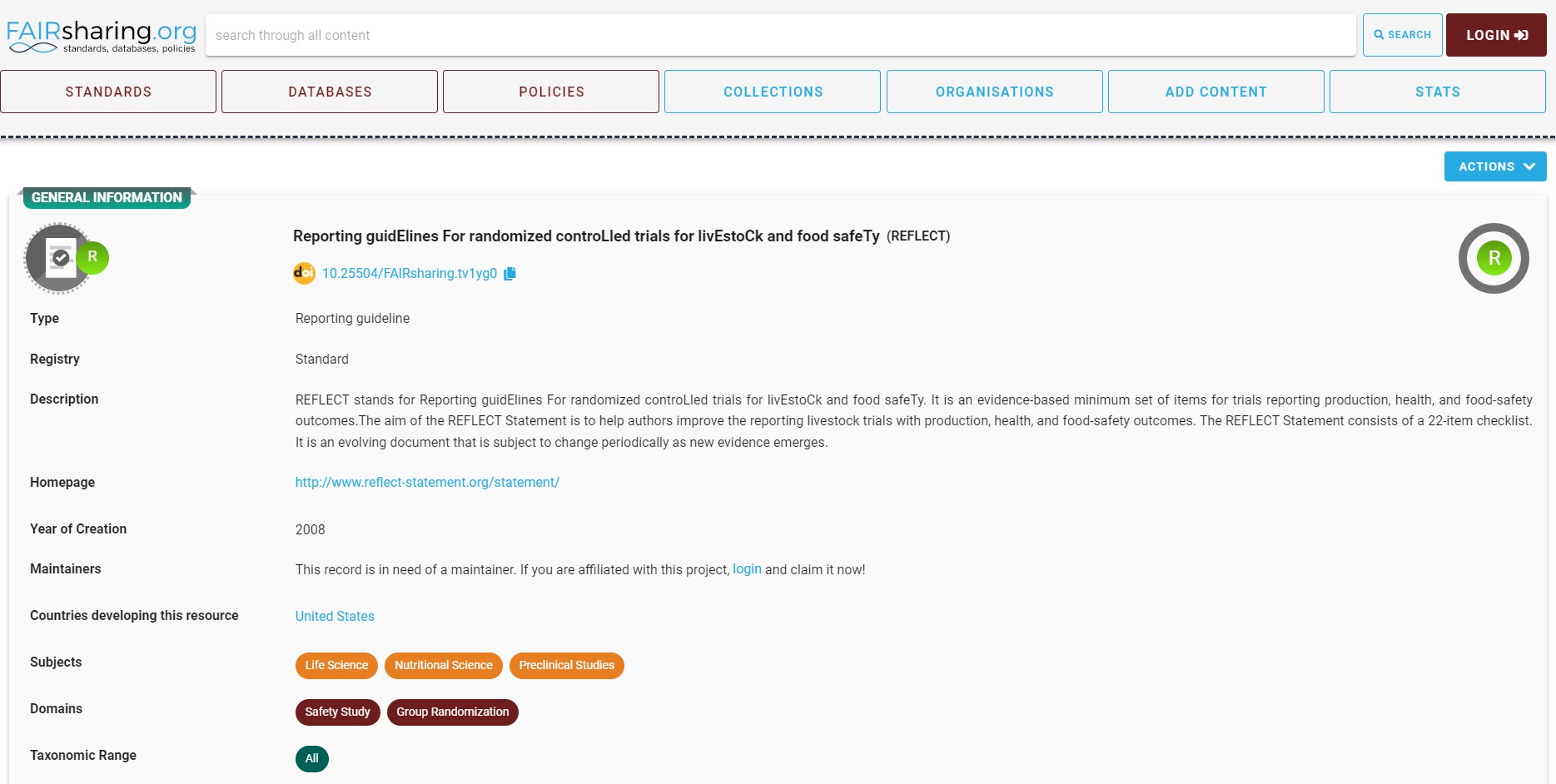
It is time to introduce you to FAIRsharing, a resource for standards, databases and policies. The FAIRsharing is an important resource for researchers to help them identify the suitable repositories, standards and databases for their data. It also contains the latest policies from from governments, funders and publishers for FAIRer data.
You can use the search wizard, to look for the guidelines for reporting the data and metadata of randomized controlled trials of the livestock and food.
In the results section, you can find REFLECT guidelines.
Following community standards
Each data type has its own community that develops guidelines to describe data appropriately and consistently. Make sure to follow the community standards when describing your data. Following standards will also make your data more reliable for other researchers, allowing it to be reused across multiple platforms. If you decide to use other guidelines outside your community, document them.
Exercise 1. Where to find your community standards
RDMkit is an open-source, community-driven site that guides life scientists to manage their research data better. This resource can be your perfect starting point for finding other tools, training materials and any recommended resources related to RDM in the life sciences.
Can you find the bioimage community standards in the RDMkit? Start here»
Solution
RDMkit covers various research data management topics and life sciences fields. You can find the community standards under the “Your domain” tab.
Inside the domain tab, you can navigate the multiple available domains with the side navigation pane. At the top, you will find “Bioimage data” tab. This page includes the following information on the bioimage community standards:
- What is bioimage data and metadata?
- Standards of bioimage research data management
- Bioimage data collection
- Data publication and archiving
Key Points
(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation (I1)
(Meta)data include qualified references to other (meta)data (A3)
Metadata are accessible, even when the data are no longer available (A2)