FAIR guiding principles
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is FAIR?
Why is FAIR important?
Pillars of FAIR
Objectives
Identify the importance of FAIR principles for the open science
Explain The difference between FAIR and open data
Contextualise the main principles of FAIR in the light of the main pillars (Identifiers, access, metadata, and registration)
Key Points
FAIR stands for Findable, Accessible, Interoperable and Reusable
What is meant by FAIRness and FAIRification of data?
Metadata, identifiers , registration, access are key components in the process of FAIRification
The FAIR Principles differ from Open data because they permit the owner of the data to control access, although as part of this they are required to define methods and instances where data could be accessed
Registration
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is a data repository?
What are types of data repositories?
Why should you upload your data to a data repository?
How to choose the right database for your dataset?
Objectives
Define what is data repository.
Illustrate the importance of indexed data repository
Summarize the steps of data indexing in a searchable repository
What is a data repository?
It is a general term used to describe any storage space you use to deposit data, metadata and any associated research. Kindly note that database is more specific and it is mainly for the storage of your data.
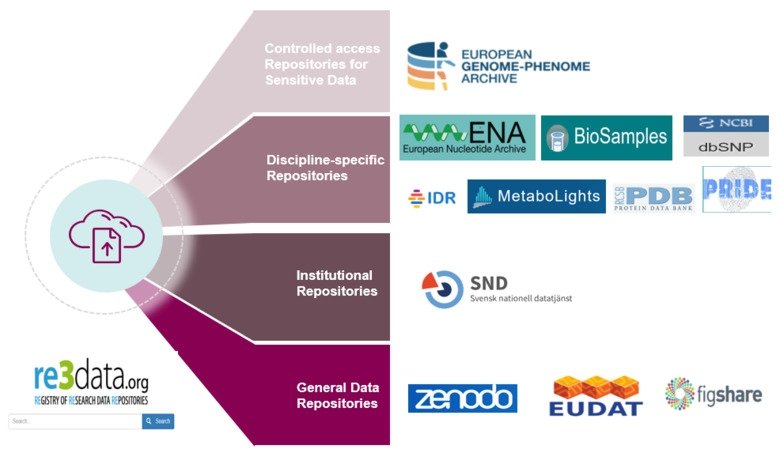
Types of data repository
Data repositories are classified based on the purpose of data repository into:
A) Controlled access repository for sensitive data: explained in details in data sharing lesson of RDMkit and we will explain this type of repository in the next episode
B) Discipline specific repository: there are known repository for different data types e.g Arrayexpress for high-throughput functional genomics experiments
C) Institutional repository: In case you can not find suitable repository for your data set, some universities have their own general purpose repositories. For instance, University of Reading Research Data Archive is a general purpose repository that have similar features e.g. controlled access … etc to other databases. It can be used for students and researchers.
D) General data repository: these are usually for data that have no public repositories e.g. Zenodo
Figure 1 summarizes these types with different examples
Why should you upload your data to a data repository?
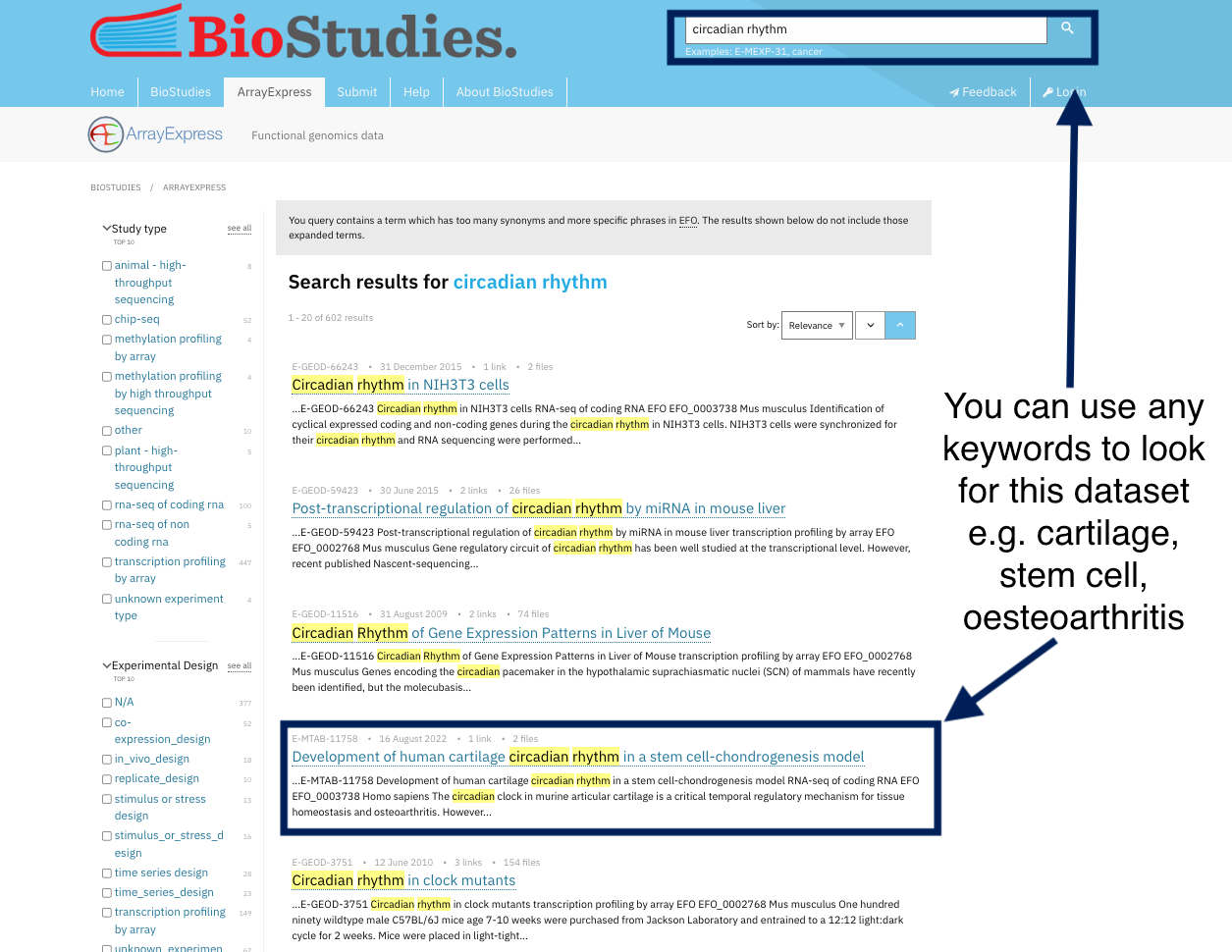
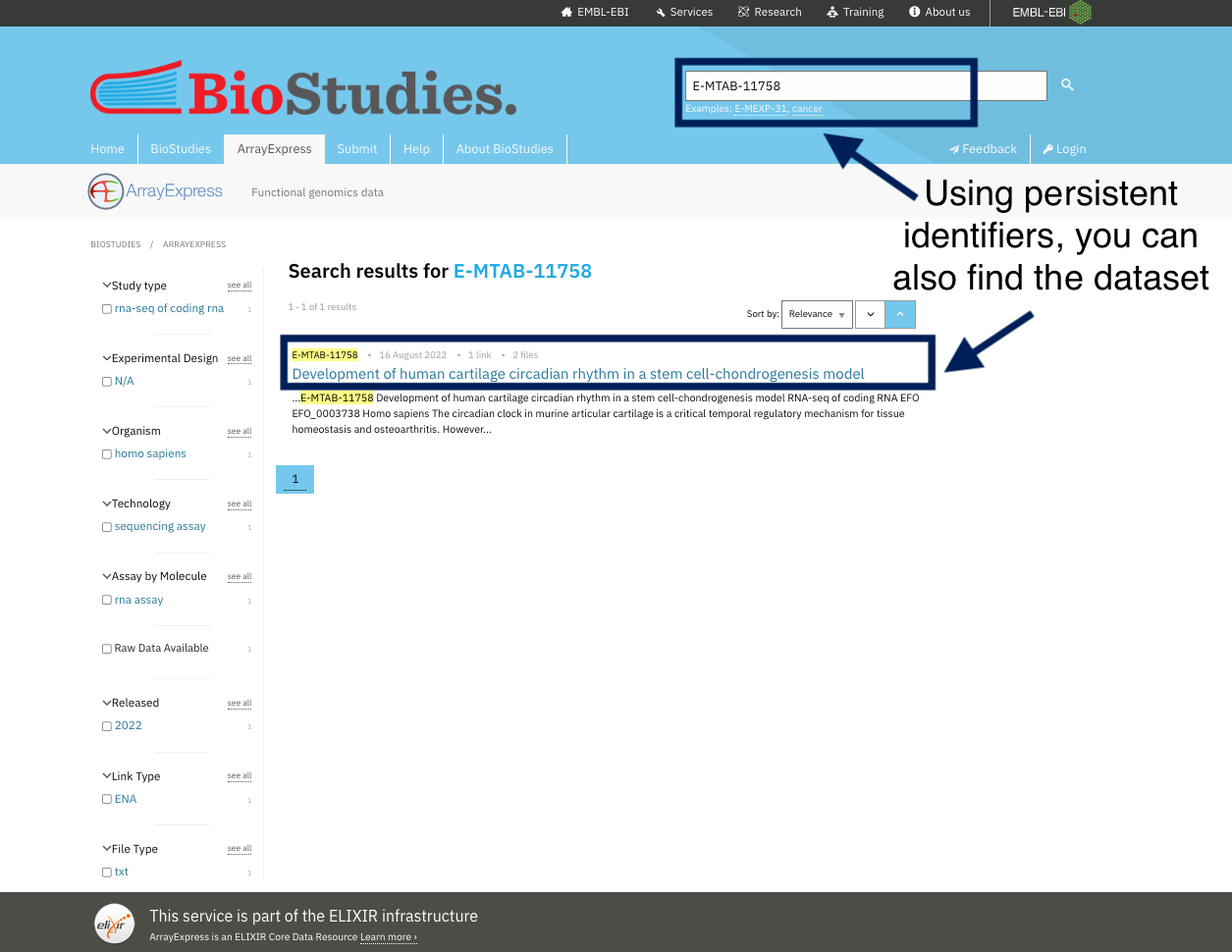
To ensure data findability, your data should be uploaded to a public repository where it can be searched and found, This will make it comply with the fourth principle of findability (F4) which states that (Meta)data are registered or indexed in a searchable resource. Examples of these databases are ArrayExpress for high-throughput functional genomics experiments. These databases have a set of rules in place to make sure that your data will be FAIR. After you upload your data into this database, they are assigned an ID and are indexed. Indexing helps researchers find your data by using persistent identifiers, keyword or even the name of researcher.
Take a look at the ArrayExpress database where all datasets are indexed, and you can simply find any dataset using the search tools. By indexing data, you can get the dataset using any keyword other than the PID. For example, if you want to locate human NSCL cell lines, you can just type this into the search toolbox, use different keywords like cartilage, stem cells and oesteoarthritis, and you will find the same dataset. Indexing and registering datasets, also means they are curated in such a way that you may discover them using different keywords.
For example, you can find the same dataset by using its identifiers or by using keywords chosen by the dataset’s authors to describe it.
Exercise 1. How to index your dataset?
One of the things you can do to index your dataset, is to upload it to Zenodo, can you use one of the resources we recommended before to know how to do this? RDMkit, FAIRcookbook, FAIRsharing
Solution
Since you want a technical guideline, FAIRcookbook and RDMkit are the best to start with. We will start with FAIRcookbook First of all, let’s understand the structure of the FAIRCookbook. For a quick overview, you can watch our RDMBites on FAIRcookbook FAIRcookbook RDMBites
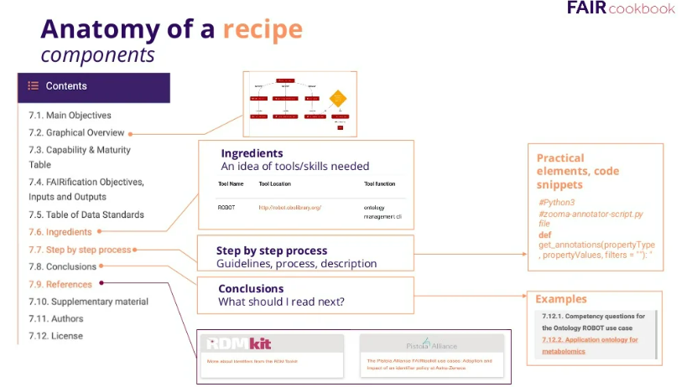
The building unit of FAIR cookbook is called a recipe, The recipe is the term used to describe instructions for how to FAIRify your data. As you see in the image, the structure of each recipe includes these main items Figure 2: 1- Graphical overview which is the mindmap for the recipe 2- Ingredients which gives you an idea for the skills needed and tools you can use to apply the recipes 3- The steps and the process 4- Recommendations of what to read next and references to your reading
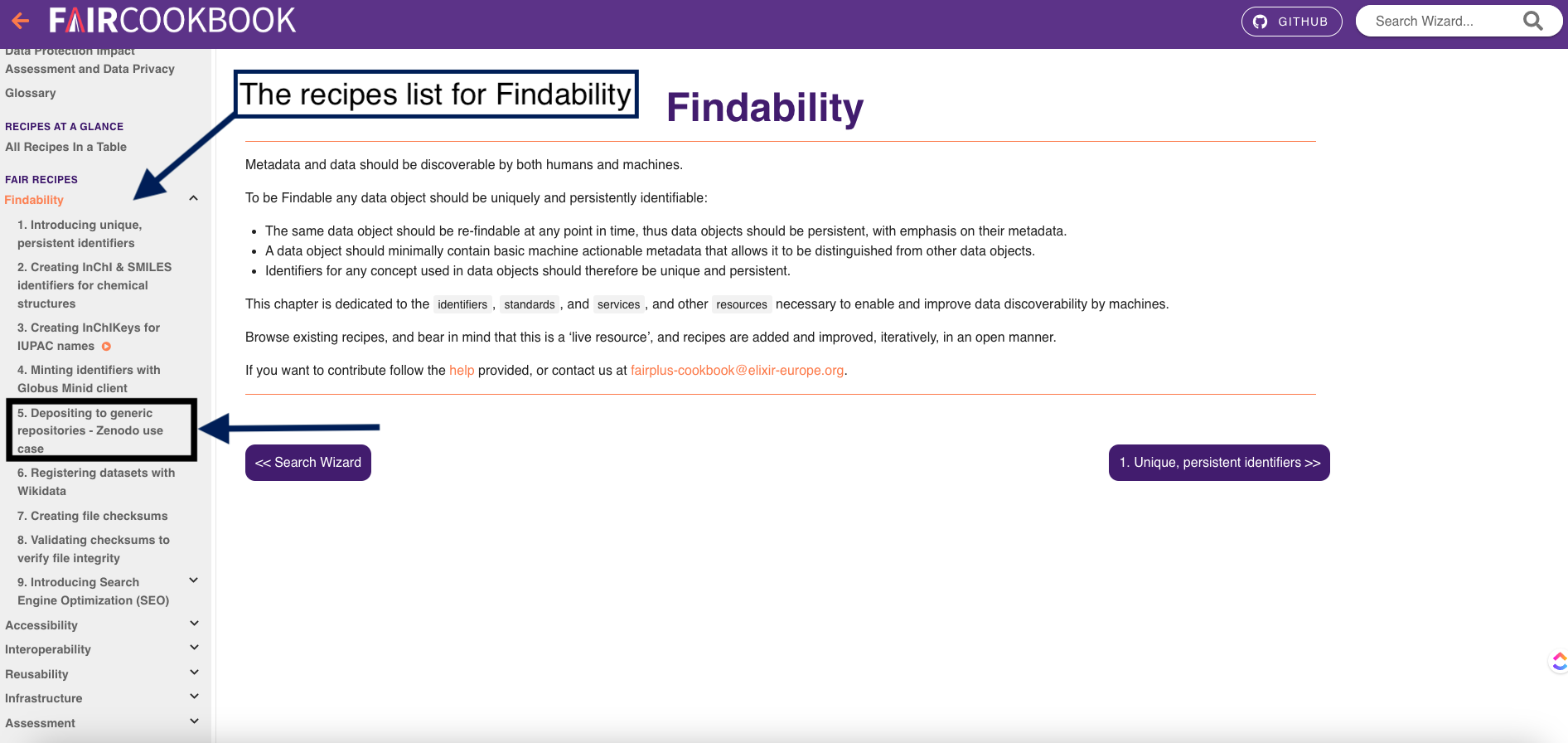
As we explained the structure of the recipe so let’s look for the suitable recipe in the FAIRcookbook So as you navigate the homepage of FAIRcookbook, you will find different tabs that covers each of FAIR principles, so for instance, if you want recipes on Accessibility of FAIR, you will find all recipes that can help you make your data accessible.
- Follow the following steps to find the recipe:
1- In this exercise, we are looking for a recipe on indexing or registering dataset in a searchable resource which you can find it in the findability tab, Can you find it in this picture?
2- Click on the findability tab
3- on the left side, you will find a navigation bar which will help you find different recipes that make your data findable.
4- As you can see here, you will find a recipe on registering datasets with Wikidata and another one on depositing to generic repositories-Zenodo use case Once you click on one of these resources, you will find the following:
A) Requirements to apply the recipe to your dataset B) The instructions C) References and further readings B) Authors and licence
In our specialized courses, we will give you examples on how to upload your data to discipline specific repository
Uploading your data to a database will make your data visible through the following:
1- Databases assign a unique persistent identifier to your data.
2- Your data will be indexed and curated, making it easier to find.
3- Some databases make it simple to connect your dataset to other datasets and link metadata to other dataset linked metadata
4- Dataset licencing, with some databases offering controlled or limited access to protect your data.
By uploading data to a database, you comply with the following FAIR principles
- F1 (Meta)data is assigned a globally unique and persistent identifier
- F3 Metadata clearly and explicitly include the identifier of the data they describe
- F4 (Meta)data is registered or indexed in a searchable resource It will also allow your data to be more accessible as the standardized communications protocol and authentication are automatically set for your data
- A1 (Meta)data is retrievable by their identifier using a standardised communications protocol
- A1.1 The protocol is open, free, and universally implementable
- A1.2 The protocol allows for an authentication and authorisation procedure, where necessary
- A2 Metadata is accessible, even when the data is no longer available
- I3 (Meta)data include qualified references to other (meta)data
- R1.1 (Meta)data is released with a clear and accessible data usage license
How to choose the right database for your dataset?
University of Reading provides an overview of the necessary criteria to choose a data repository. We can summarize it in the following bullet points:
- Check funders recommendations It is always better to upload your data to funders recommendied data repositories. For instance, Biotechnology and Biological Sciences Research Council (BBSRC) funds and recommend many databases including European Bioinformatics Institute
- Publishers Publishers prefers discipline specific repository, check guidelines before you submit your manuscript.
- Community standards Check the community standards for your data, you can find more information RDMkit guidelines
- If you still cannot find the right one for you, look for resources that describe the databases and check if it fits your data, you might consider the following:
A) Accessibility options
B) Licence
- One of resources that can help you is FAIRsharing, it provides a registry for different databases and repositories. Here is an example where the FAIR sharing provides you with information regarding protein database. It has the following information
- General information
- Which policies use this database?
- Related community standards
- Organization maintaining this database
- Documentation and support
- Licence
Exercise 1. How to choose the right dataset?
You are a researcher in plant sciences and want to know what are the available databases for plant genomes?
Solution
It is the time to introduce you to FAIRsharing, an important resource for metadata standards, databases and policies. The FAIRsharing is an important resource for researchers to help them identify the suitable repositories, standards and databases for their data. It also contains the latest policies from from governments, funders and publishers for FAIRer data. In the following short video, you can find that plant ensembl is the one you can use for the plant genes
Resources
Our resources provide an overview of data repositories and examples
The FAIR cookbook and RDMkit both provide excellent instructions for uploading your data into databases:
- FAIRcookbook recipe on Depositing to generic repositories- Zenodo use
- FAIRcookbook recipe on Registering Datasets in Wikidata
- RDMkit guidelines on Data publications and depostion
- RDMkit guidelines on Finding and reusing existing data
- FAIRcookbook recipe on Search engine optimization
- FAIRsharing offers a nice portal to different examples of databases
Key Points
{“This episode covers the following FAIR principles”=>nil}
(Meta)data are registered or indexed in a searchable resource (F4)
(Meta)data are released with a clear and accessible data usage licence (R1.1)
Access
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is protocol and authentication?
What are the types of transfer protocols?
What is data usage licence?
What is sensitive data?
Objectives
To illustrate what is the communications protocol and the criteria for open and free protocol
To give examples of databases that uses a protocol with different authentication process
To interpret the usage licence associated with different data sets
Access to the data
As a researcher, when you plan your research project, you have to determine who can have access to your data, how you will provide the access and under what condition. You do not need to wait after data collection to start thinking about the access to your data. To start writing your acces plan, first we will explain to you what are different types of data access, what is the best for you and how to write your data access plan
Types of access
there are four types of data access as explained by RDMkit:
- Open access: Any one can access the data
- Registered access or authentication procedure: researchers have to register and go through process of authentication to have the right to access the data
- Controlled access or Data Access Committees (DACs): researchers who wants access to your data will apply and their application will be reviewed by a data access committee who will assess the application and make sure that resaerchers will abide by the criteria you have specified
- Access upon request (not recommended): you provide your contact details for any researcher who can contact you to get access to your data. Contact details should be provided in the metadata which is made publicly available.
To get more detailed explanation, check RDMkit explanation of data sharing
Exercise
Imagine you are a principle investigator writing data access plan, what are the factors you need to consider to determine the type of access for your data?
Solution
To know what to write for data access plan, you can use a tool called “Data stewardship wizard” that provides guidelines on the process of writing DMP for your research project. It has a full chapter on writing access plan. Let’s first explain what is data stewardship wizard
What is Data stewardship wizard (DSW)?
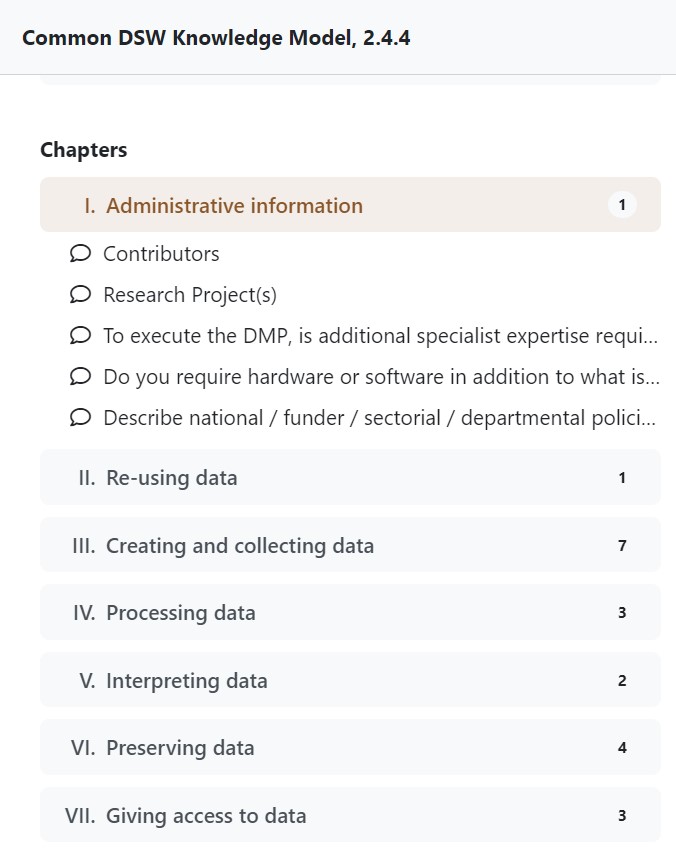
It is one of ELIXIR resources which helps you create your DMP. It uses a knowledge model, which includes information about what questions to ask » and how to ask them based on the needs of the research field or organisation. The knowledge model covers seven chapters:
- Administrative information
- Re-using data
- Creating and collecting data
- Processing data
- Interpreting data
- Preserving data
- Giving access to the data
For each of these topics, we have a set of questions that helps you write your DMP. For each question, there are tags that help you know funding bodies questions (Figure 2)
After introducing the DSW, let’s answer the following question, How to choose the right data access plan for you? Based on DSW, there are main four questions
- Will you be working with the philosophy ‘as open as possible’ for your data?
- Can all of your data become completely open over time?
- Will you use temporary restrictions on the reuse of the data (embargo)?
- Will metadata be available openly? For each of these questions, there is a follow-up questions based on your answer to the main question
One of things you have to consider in addition to the above criteria, is to determine the data usage licence for your data
Data usage licence
This describes the legal rights on how others use your data. As you publish your data, you should describe clearly in what capacity your data can be used. Bear in mind that description of licence is important to allow machine and human reusability of your data. There are many licence that can be used e.g. MIT licence or Common creative licence. These licences provide accurate description of the rights of data reuse, Please have a look at resources in the description box to know more about these licences.
 {alt=’alt text’}
{alt=’alt text’}
Exercise
- From the this RDMkit guideline on types of licence, what is the licence used by the following datasets: 1- A large-scale COVID-19 Twitter chatter dataset for open scientific research - an international > collaboration 2- RNA-seq of circadian timeseries sampling (LL2-3) of 13-14 day old Arabidopsis thaliana Col-0 (24 h to 68 > h, sampled every 4 h)
Solution
The link we provided, provided a nice explanation on types of licence and as you read the following section, you will find the following part:
{alt=’alt text’} From this section, you can clearly understand the type of licence used for: 1- A large-scale COVID-19 Twitter chatter dataset for open scientific research - an international collaboration is CC-BY-4 2- RNA-seq of circadian timeseries sampling (LL2-3) of 13-14 day old Arabidopsis thaliana Col-0 (24 h to 68 > h, sampled every 4 h) is CC-BY-4
As you are uploading your data to a data repository, the following definitions are important for you to understand the type of access. Communication protocol and authentication are used by different databases to protect your data and control access to your data
Standard communication protocol
Simply put, a protocol is a method that connects two computers, the protocol ensure security, and authenticity of your data. Once the safety and authenticity of the data is verified, the transfer of data to another computer happens.
Having a protocol does not guarantee that your data are accessible. However, you can choose a protocol that is free, open and allow easy and exchange of information. One of the steps you can do is to choose the right database, so when you upload your data into database, the database executes a protocol that allows the user to load data in the user’s web browser. This protocol allows the easy access of the data but still secures the data.
Authentication process
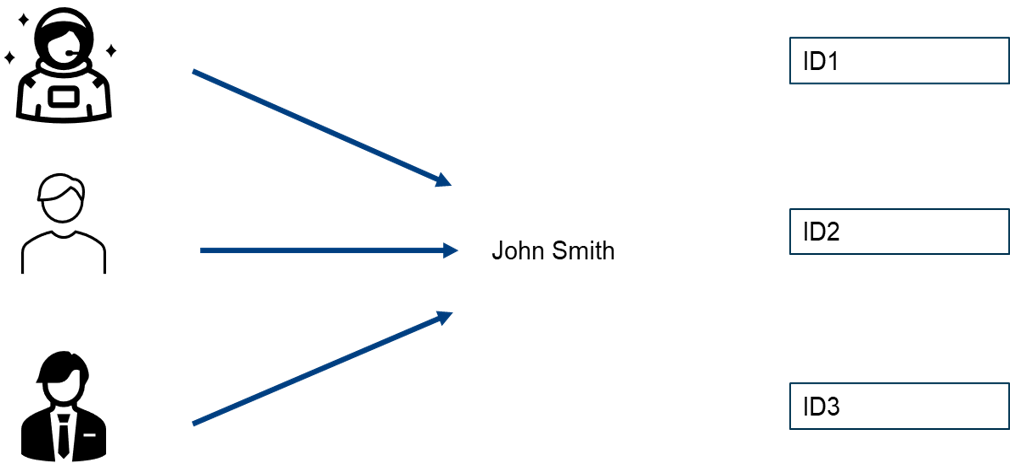
It is the process that a protocol uses for verification. To know what authentication is, suppose we have three people named John Smith. We do not know which one submitted the data. This is through assigning a unique ID for each one that is interpreted by machines and humans so you would know who is the actual person that submitted the data. Doing so is a form of authentication and this is used by many databases like Zenodo, where you can sign-up using ORCID-ID allowing the database to identify you.
Exercise
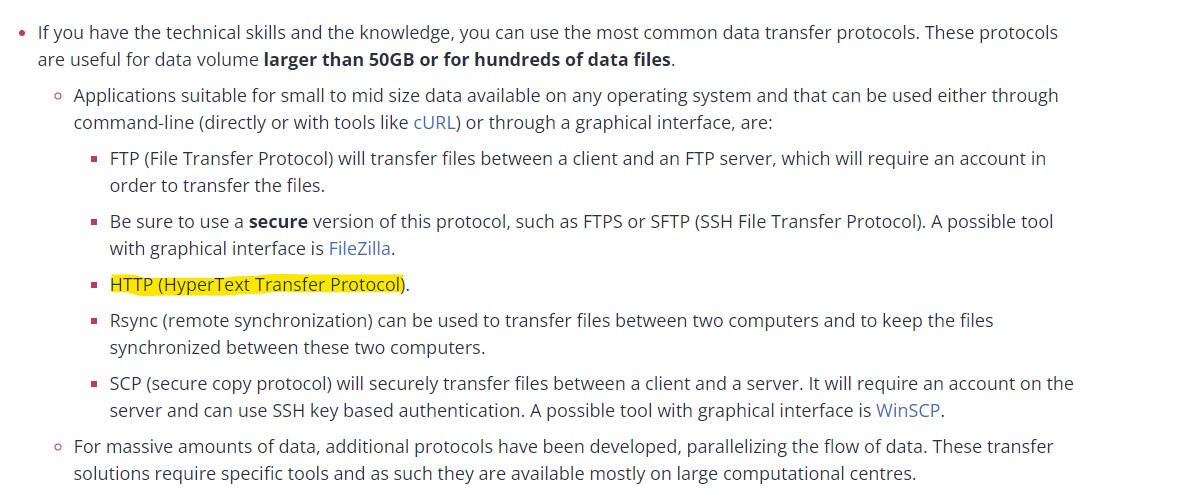
After reading RDMkit guidelines on different protocol types, do you know what is the protocol used in arrayexpress?
Solution
As we explained before on how to use the RDMkit, going through the Protocols and safety of data transfer, you will find different types of protocols explained
From this part, you can understand that the protocol used for the arrayexpress is HTTP (HyperText Transfer Protocol) (highlighted in yellow) » in the following figure
Sensitive data
Sensitive data are type of data that, if made publicly available, could cause consequences for individuals, groups, nations, or ecosystems and need to be secured from unauthorised access. To determine whether your data is sensitive, you should consult national laws, which vary by country.
If your data is following this definition, you have to de-identify your data. Data deidentification is a process through which data cannot be identified through the study team nor the users of the data.
- It includes two processes:
1- Data anonymization
2- Data Pseudonymization
Exercise
- From the this RDMkit guideline on Sensitive data, can you out find what is the data anonymization and data pseudonymization?
Solution
The link we provided, provided a nice explanation on sensitive data and as you read the following section from RDMKit, definitions are the following: Data anonymization is the process of irreversibly modifying personal data in such a way that subjects cannot be identified directly or indirectly by anyone, including the study team. If data are anonymized, no > one can link data back to the subject.
Pseudonymization is a process where identifying-fields within data records are replaced by artificial identifiers called pseudonyms or pseudonymized IDs. Pseudonymization ensures no one can link data back to the subject, apart from nominated members of the study team who will be able to link pseudonyms to identifying > records, such as name and address.
If you are working with sensitive data, you have to declare the data permitted uses before using the data. In addition, if you are writing data management plan (DMP), you will have to mention the following:
- How will you collect the data?
- How is Data annonymization and pseudonymization handled?
To know what are the other steps you need to do and write, you can check DSW For sensitive data, the DSW have questions dedicated to describing the collection and processing of sensitive data.
Resources
- A nice recipe from FAIRcookbook on SSH protocols
- A nice explanation from RDMkit on protocols and how they will help you protect your dataProtocols and safety of data transfer
Having your work licenced does not sound simple as it seems; here are some resources to help you find the > correct licence for you:
- Why should you assign licence to your protocol from RDMkit Licencing
- A nice recipe from FAIRcookbook with step-by-step instructions for
- licence
- software licence
- Data licence
- Declaring data permitted uses
- To know more about creative common licence, check this link Creative commons licence
To get more information on sensitive data, you can have a look on these reources:
Key Points
{“This episode covers the following FAIR principles”=>nil}
(A1) (meta)data are retrievable by their identifier using a standardised communications protocol
(R1.1) meta(data) are released with a clear and accessible data usage licence
Persistent identifiers
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is a persistant identifiers?
What is the structure of identifiers?
Why it is important for your dataset to have an identifiers?
Objectives
Explain the definition and importance of using identifiers
Illustrate what are the persistent identifiers
Give examples of the structure of persistent identifiers
Persistent identifiers
Identifiers are a long-lasting references to a digital resources such as datasets, metadata .. etc. They provide the information required to reliably identify, verify and locate your research data. Commonly, a persistent identifier is a unique record ID in a database, or a unique URL that takes a researcher to the data in question, in a database.
That resource might be a publication, dataset, or person. Persistent identifiers have to be unique, globally only your data are identified by this ID that is never used by anyone in the whole world. In addition, these IDs and must not do not become invalid over time. Watch our RDMbBites on the persistent identifiers to understand more.
Identifiers are very important concept of the FAIR principle. They are considered one of the pillars for the FAIR principles. It makes your data more Findable (F)
It is important to note that when you upload your data to a public repository, the repository will create this ID for you automatically.
Based on how to FAIR, there are many resources that can help you know which databases will assign PID to your data. One of these resources is FAIR sharing, it provides you with a list of databases grouped by domains and organizations.
The Structure of persistent identifiers
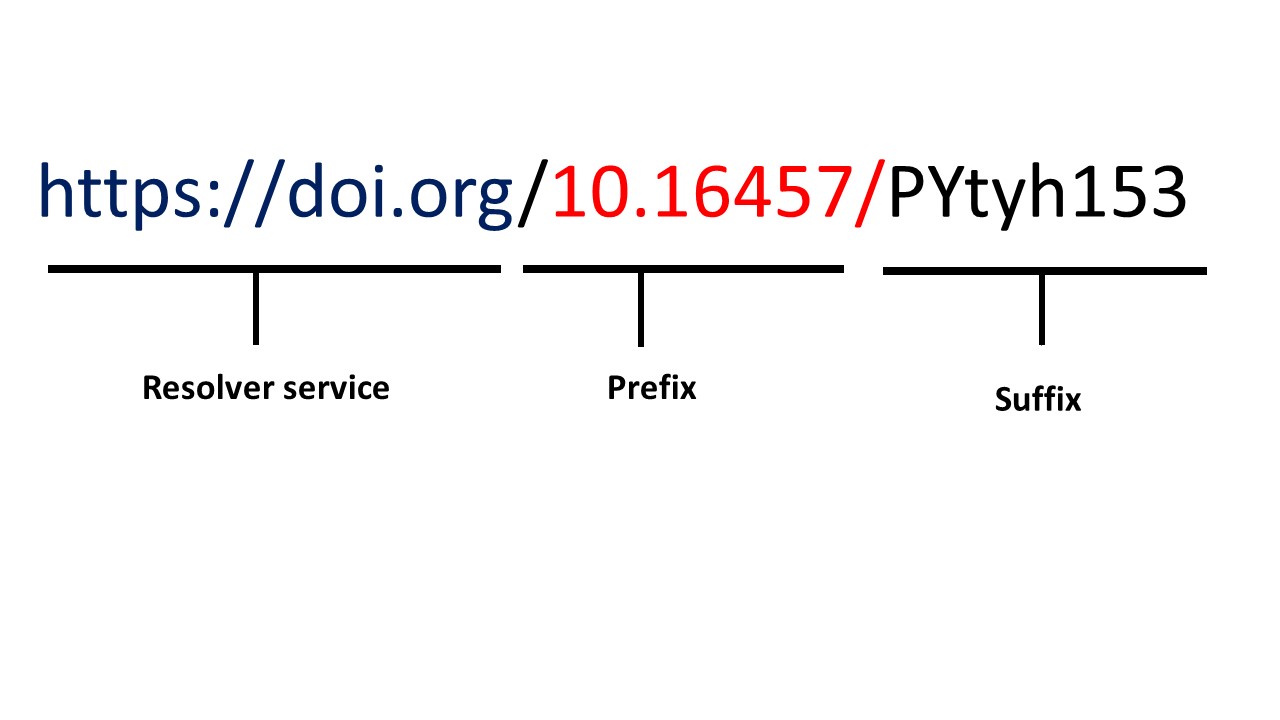
As you can see in this picture, the structure of any identifiers consist of
- The initial resolver service: domain which is unique and specific to each community e.g. ORCID for researchers
- Prefix: Unique number that represent category e.g. for DOI specific numbers refer to the publisher and directory
- Suffix: The unique dataset number and it is unique under its prefix
Exercise 1. Find the PID
From FAIRsharing, can you find the right database for protein dataset and explore its PID structure?
Solution
If you follow the steps in the following screen recording, you will find plant genomics and phenotypes. In this database, all datasets are assigned digital object identifier (DOI)
The DOI is a persisitent identifiers that follows the structure we explained before
Resources
The resources listed below provide an overview of the information you need to know about identifiers.
Unique and persistent identifiers: this link provide a nice and practical explanation of the unique and persistent identifiers > from FAIRCookbook
Identifiers: another nice explanation from RDMkit
Machine actionability: identifiers are also important for machine readability, a nice explanation from RDMkit that describes machine readability
Examples and explanation of different identifiers from FAIRsharing.org https://fairsharing.org/search? recordType=identifier_schema
Key Points
(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation (I1)
(Meta)data include qualified references to other (meta)data (A3)
Metadata are accessible, even when the data are no longer available (A2)
Metadata
Overview
Teaching: 40 min
Exercises: 10 minQuestions
What is FAIR? What is the origin of the FAIR movement?
Why is FAIR important?
What is the difference between FAIRness and FAIRification of data?
Objectives
Define metadata and its various types
Recall the community standards and how to apply them to data and metadata
What is metadata?
Metadata refers to the information that describes your data.
In other words, imagine you have an Excel spreadsheet containing data values for an assay. You would use column headings to assign meaning and context. These column headings are your metadata, explaining the data values in each cell. In addition, any documentation or explanation of the accompanying excel file is also considered metadata.
Let’s look at Figure 1, showcasing a spreadsheet containing data for a clinical assay. In this example, the data are the patient ID, disease type, and heart rate values. The metadata, the column headings, describe that those values correspond to the patient ID, disease type, and heart rate, as well as the name of the cohort and the contact e-mail.

What information could we add to better understand the data contained in the dataset?
We could add additional metadata to indicate data provenance, i.e. data origin, what happens to it or where it moves over time. In this case, we should add more information about the cohort name. “Human Welsh Cohort” does not tell us much about the data if compared to other Welsh cohorts. Instead, we could include the following:
- A unique ID or detailed title for the cohort
- A project URL describing its origin and composition At first glance, the column headings seem descriptive. However, taking a closer look, we can see that the “disease type” column captures both the disease type and stage, which can cause ambiguity:
- In row 3, it is unclear whether the disease is diabetes mellitus or insipidus
- In row 4, it is unclear whether the type of diabetes mellitus is 1 or 2
- There is an empty space in the final row. It is unclear whether this is due to a lack of information or that the patient does not have diabetes
Instead, we can create two separate columns: one for disease type and one for the stage (see Figure 2).

The importance of metadata
Metadata is small and can be easily maintained not only in the database but in personal computers. The maintenance of datasets in a public database comes at a cost. It can be minimised when maintaining the metadata instead. In addition, metadata is also highly efficient for sharing sensitive data. The details available are those provided in the metadata, such contact details of the researchers, how to get the data and how it was generated.
Types of metadata
We’ve seen that metadata can describe various aspects of your dataset. Based on how to FAIR, there are three types of metadata:
- Descriptive Metadata: this metadata type help you identify the dataset e.g identifiers, publication name .. etc
- Structural Metadata describes how the dataset is generated and structured internally e.g. analysis, units of measurements, collection methods … etc
- Administrative metadata describes who was in charge of the data, who worked on the project, and how much money was spent.
Let’s look at an example using microarray data from the ArrayExpress database (Figure 3) to locate the different types of metadata that we have defined.
 We can observe:
We can observe:
- Administrative metadata: authors and organisations underneath the dataset title, and the information in “Publication”
- Descriptive metadata: “Description” section that sumarises the information contained in the dataset
- Structural Metadata: “Protocols”, “Samples” and “Assay and Data” sections describing the structure of the dataset and how it was generated
Metadata is also a documentation of data lifecycle. Through metadata, you describe what happened to metadata at each step of data life cycle. We use the term Data provenance to describe these steps. Provenance is the detailed description of the history of the data and how it is generated. Here is an example from arrayexpress database where there is accurate description of the microarray life cycle. As you can see in this example from E-MTAB-6980 dataset, there is rich description of the study design, organism, platform and timing of data collection.
How to use metadata to describe your dataset?
Metadata is data about data! It is important to know how to document it and use the right vocabulaeries to make your metadata FAIR. Using the right voacabularies will help standardize the way we describe our data.
Vocabularies and ontologies Controlled vocabularies: are list of terms that describes certain domain of knowledge. Vocabularies usually include definition of the term and any synonyms. For instance, medical subject headings (MeSH) terms is a common resource for controlled vocabularies. For instance, you can describe carotid artery as “common carotid artery” or “carotid sinus”
When you describe your data, you also need to describe the relationship between different vocabularies, which we call it ontologies
Ontologies: describe the relationship between different terms. There are many resources that you can use to get ontologies for your metadata. BRENDA, an ELIXIR resource that helps you get the right ontologies for your metadata.
Exercise
You are researcher working in the field of food safety and you are doing clinical trial, do you know how to > choose the right vocabularies and ontologies for it?
solution
It is time to introduce you to FAIRsharing, a resource for standards, databases and policies. The FAIRsharing is an important resource for researchers to help them identify the suitable repositories, standards and databases for their data. It also contains the latest policies from from governments, funders and publishers for FAIRer data.
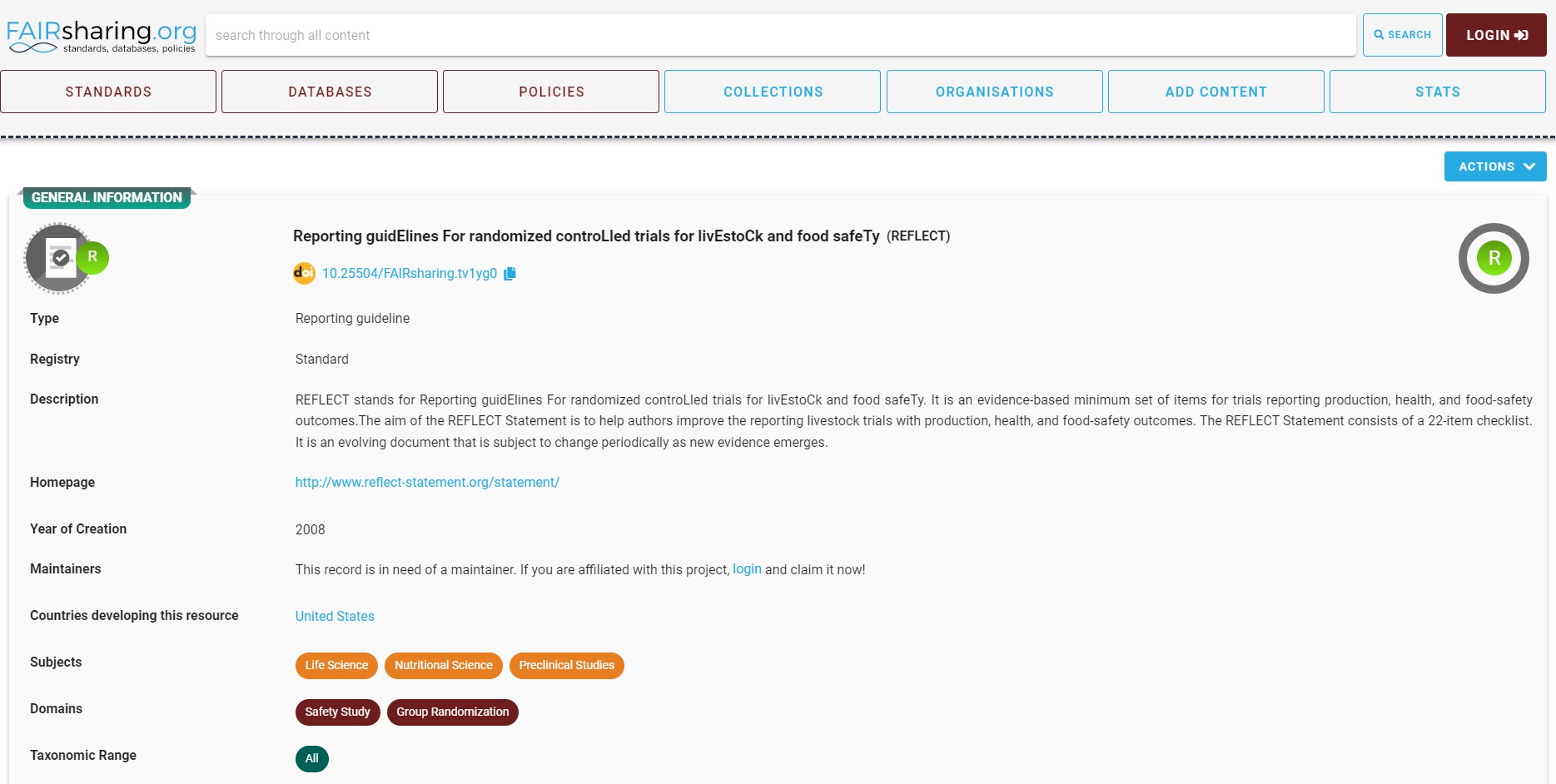
You can use the search wizard, to look for the guidelines for reporting the data and metadata of randomized controlled trials of the livestock and food.
In the results section, you can find REFLECT guidelines.
Following community standards
Each data type has its own community that develops guidelines to describe data appropriately and consistently. Make sure to follow the community standards when describing your data. Following standards will also make your data more reliable for other researchers, allowing it to be reused across multiple platforms. If you decide to use other guidelines outside your community, document them.
Exercise 1. Where to find your community standards
RDMkit is an open-source, community-driven site that guides life scientists to manage their research data better. This resource can be your perfect starting point for finding other tools, training materials and any recommended resources related to RDM in the life sciences.
Can you find the bioimage community standards in the RDMkit? Start here»
Solution
RDMkit covers various research data management topics and life sciences fields. You can find the community standards under the “Your domain” tab.
Inside the domain tab, you can navigate the multiple available domains with the side navigation pane. At the top, you will find “Bioimage data” tab. This page includes the following information on the bioimage community standards:
- What is bioimage data and metadata?
- Standards of bioimage research data management
- Bioimage data collection
- Data publication and archiving
Key Points
(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation (I1)
(Meta)data include qualified references to other (meta)data (A3)
Metadata are accessible, even when the data are no longer available (A2)